New Autism Subtypes Identified Based on Genes, Individual Traits
- Published11 Nov 2025
- Author Kristel Tjandra
- Source BrainFacts/SfN

As a spectrum disorder, autism presents in myriad and complex ways. Now, researchers have discovered four distinct subtypes of autism, each with its own genetic profile. The discovery could unravel the mystery of what causes the neurodevelopmental disorder and advance efforts to personalize interventions for it.
“We have this analogy of trying to put together a puzzle, but actually, you have four different puzzles mixed together, and you can’t find any common pieces,” says Natalie Sauerwald, a computational biologist at the Flatiron Institute in New York.
Sauerwald is one of the lead authors of a study published in July 2025 in Nature Genetics, where scientists assigned individuals with ASD into four subtypes based on their observable behaviors and then analyzed their genetic differences. Funded by the Simons Foundation and the U.S. National Institute of Health, the study provides a new framework for understanding the diversity of autism.
While environmental factors likely play some role in the risk of autism, the vast majority of autism cases have genetic root causes, said Maria Chahrour, a neurogeneticist at the University of Texas Southwestern Medical Center, who was not involved in the study. Chahrour emphasized that does not mean autism must be passed down from the parents; instead, genetic mutations in autism often occur spontaneously (also known as de novo) during reproductive cell division.
“We know that it's not one autism; there are many autisms,” said Chahrour. “Genetic variants in autism are almost always private.” In other words, the specific change in the DNA sequence of a person diagnosed with autism can look entirely different from that of another person with the disorder.
Pamela Feliciano, a neurodevelopmental geneticist at the Boston Children’s Hospital and Harvard Medical School, says achieving a strong understanding of different genetic pathways underlying different categories of autism is going to take a lot of data and time. Feliciano directs SPARK, the largest autism research study in the world, from which Sauerwald and colleagues derived data for the new study.
Since 2016, researchers behind the SPARK study have collected data from more than 380,000 individuals in the U.S. who were either diagnosed or have a family member with autism. SPARK participants fill out questionnaires on cognitive and behavioral traits, medical and family histories, and submit saliva samples for DNA analysis to contribute to autism research.
A dataset combining the genetics and traits associated with autism “is extremely rare” at this scale said Aviya Litman, a graduate student at Princeton University and co-lead author of the study.
Even though scientists have identified hundreds of genes associated with autism, each of these genes accounts for a very small proportion of autism cases, making the disorder highly heterogeneous. “You can think of it [autism] almost like a collection of individual rare diseases,” Chahrour said.
To make sense of this diversity, scientists have tried to group different autism-associated genes and traits into categories. The new study by Litman, Sauerwald, and colleagues shows many of the clinical attributes observed in people with autism can be mapped to distinct genetic and biological mechanisms. Chahrour believes the study offers opportunities for testing new hypotheses about how researchers understand the biology of autism.
Sifting Through a Large Dataset for Patterns
For the study, Litman, Sauerwald, and colleagues parsed through data from 5,392 autistic individuals aged four to 18 and their neurotypical siblings who participated in the SPARK study.
“We all have some level of mutations that were not in our parents, that just occurred through cell division before we were born, and those are very heavily implicated in autism,” Sauerwald said. “Having siblings with very similar genetics, but different sets of de novo mutations, is really powerful to study.”
Using a statistical model, the researchers investigated 239 of the reported autism-related traits and assigned them into previously established clinical criteria or core autism features, such as disruptive behavior, communication deficit, developmental delays (DD), and anxiety or mood symptoms. This allowed them to describe any patterns in the traits more easily.
“From the beginning, our approach has been phenotype first,” Litman said. This means the researchers based their grouping on observable traits rather than an individual’s genetic makeup.
This approach differs from other studies linking autism-associated genetic variation to individual’s traits. Since one trait can influence another, the researchers anticipated that analyzing co-occurring traits would reveal patterns better reflecting the experience of the whole person.
Four Subtypes Align with Genetic Patterns
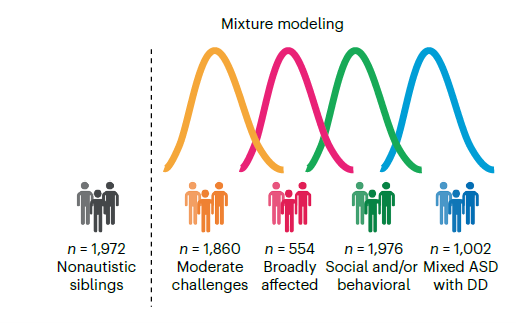
From their analysis, the researchers identified four possible classes or subtypes based on the probability of having a combination of autism-related traits:
- The “Social and/or behavioral” subtype consisted of individuals who scored high on many of the core autism features and exhibited traits of co-occurring conditions such as mood and attention disorders but show no signs of developmental delays.
- The “Moderate challenges” subtype included those who scored below other autistic participants across all the core autism features.
- The “Broadly affected” subtype consisted of those who are more affected by all of the core autism criteria than others, and high levels of co-occurring conditions.
- The “Mixed Autism Spectrum Disorder (ASD) with DD” subtype included individuals who showed some core social communication challenges, developmental delays, and restricted or repetitive behaviors but tend not to have mood disorder, attention challenges, or disruptive behavior.
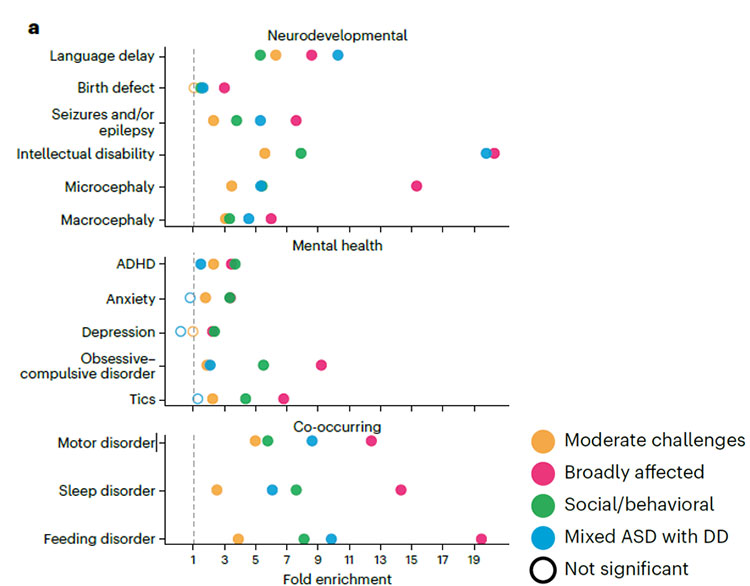
Individuals in the Broadly affected and Mixed ASD with DD subtypes tend to get diagnosed earlier in life than others due to more apparent developmental delays.
When weighed against the genetic changes in the individuals, the researchers found distinct genetic signal patterns matching the characteristics of these four classes of trait patterns.
For instance, they found de novo variants (mutations that are not inherited from either parent) associated with fragile X syndrome, where individuals experience developmental delays and intellectual disabilities, were most strongly associated with the Broadly affected subtype. On the other hand, the Social/behavioral subtype had the highest genetic signals associated with attention-deficit/hyperactivity disorder (ADHD) and depression compared to the other subtypes.
While the study looked at four classes of trait patterns in the data, “it doesn't mean that there are only four classes of autism,” Litman said. The current study is limited to the number of data points and the demographics of the participants. “It's very important to go beyond this, to expand the dataset, to look at a wider diversity of individuals,” she added.
Chahrour also found this to be true. In a study she led involving individuals with ASD from ancestrally diverse backgrounds, she found certain genetic variations can occur at different frequencies depending on a person’s ancestry.
For example, her group has identified certain gene variants associated with autism in East African individuals that have never been reported elsewhere. These variants may be missed in the current study, since a significant proportion of the SPARK participants are of European descent. “When you're interpreting results based on data collected from one ancestry, that may not necessarily apply to other ancestries,” she said.
In addition, as they grow, the young individuals in the cohort may experience changes in certain traits depending on factors such as the kind of therapy they receive. Because of these reasons, it is important to expand the diversity of the cohort in future studies, Chahrour said.
“Understanding the heterogeneity in autism has been a really challenging problem for biologists, for a long time,” Feliciano said. While the study will need to be replicated and validated for clinical application, “this is a first step towards being able to have tools in the clinic that are helpful to families.
CONTENT PROVIDED BY
BrainFacts/SfN
References
Feliciano, P., Daniels, A. M., Snyder, L. G., Beaumont, A., Camba, A., Esler, A., Gulsrud, A. G., Mason, A., Gutierrez, A., Nicholson, A., Paolicelli, A. M., McKenzie, A. P., Rachubinski, A. L., Stephens, A. N., Simon, A. R., Stedman, A., Shocklee, A. D., Swanson, A., Finucane, B., . . . Chung, W. K. (2018). SPARK: A US cohort of 50,000 families to accelerate autism research. Neuron, 97(3), 488–493. https://doi.org/10.1016/j.neuron.2018.01.015
Gogate, A., Kaur, K., Khalil, R., Bashtawi, M., Morris, M. A., Goodspeed, K., Evans, P., & Chahrour, M. H. (2024). The genetic landscape of autism spectrum disorder in an ancestrally diverse cohort. Npj Genomic Medicine, 9(1). https://doi.org/10.1038/s41525-024-00444-6
Harris, J. C. (2019). The necessity to identify subtypes of autism spectrum disorder. JAMA Psychiatry, 76(11), 1116. https://doi.org/10.1001/jamapsychiatry.2019.1928
J., Ellis, C., Oakley, B., Loth, E., Charman, T., Murphy, D., . . . Baron-Cohen, S. (2022). Genetic correlates of phenotypic heterogeneity in autism. Nature Genetics, 54(9), 1293–1304. https://doi.org/10.1038/s41588-022-01072-5
Litman, A., Sauerwald, N., Snyder, L. G., Foss-Feig, J., Park, C. Y., Hao, Y., Dinstein, I., Theesfeld, C. L., & Troyanskaya, O. G. (2025). Decomposition of phenotypic heterogeneity in autism reveals underlying genetic programs. Nature Genetics. https://doi.org/10.1038/s41588-025-02224-z
Tuncay, I. O., DeVries, D., Gogate, A., Kaur, K., Kumar, A., Xing, C., Goodspeed, K., Seyoum-Tesfa, L., & Chahrour, M. H. (2023). The genetics of autism spectrum disorder in an East African familial cohort. Cell Genomics, 3(7), 100322. https://doi.org/10.1016/j.xgen.2023.100322
Warrier, V., Zhang, X., Reed, P., Havdahl, A., Moore, T. M., Cliquet, F., Leblond, C. S., Rolland, T., Rosengren, A., Caceres, A. S. J., Hayward, H., Crawley, D., Faulkner, J., Sabet, J., Ellis, C., Oakley, B., Loth, E., Charman, T., Murphy, D., . . . Baron-Cohen, S. (2022). Genetic correlates of phenotypic heterogeneity in autism. Nature Genetics, 54(9), 1293–1304. https://doi.org/10.1038/s41588-022-01072-5
What to Read Next
Also In Childhood Disorders
Trending
Popular articles on BrainFacts.org