Insight Into Human Behavior Through Animal Models
- Reviewed6 Mar 2023
- Author Susan Rojahn
- Source BrainFacts/SfN

Our behaviors are what we as living organisms, do. And peering inside our brains to understand how we do what we do can be illuminating. So to understand how brain function drives behaviors in humans, researchers often turn to animal models.
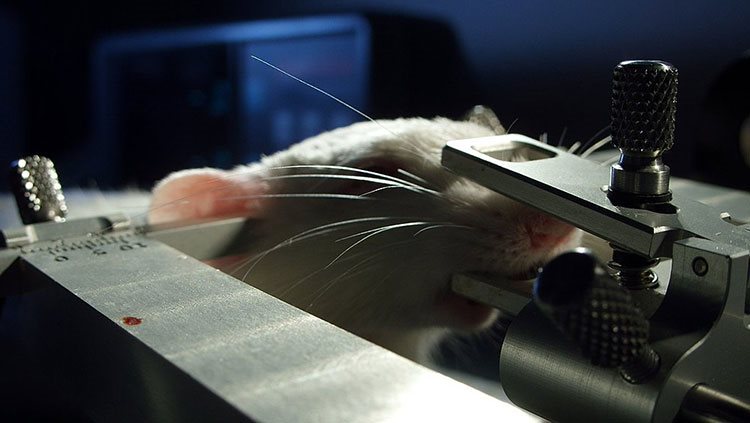
An eight-inch-long marine slug may not look like a very promising model of brain function, but over the years, the animal known as Aplysia has helped scientists uncover many principles of learning and memory. Aplysia has relatively few neurons (around 10,000, compared to approximately 86 billion in humans), but some of its neurons are large enough to be seen with the naked eye. Aplysia also exhibits simple behaviors that can be modified with training. For example, Aplysia will reflexively withdraw its gill after receiving an electric shock to its tail. It can be trained to withdraw its gill in response to an innocuous touch which, during training, was paired with an electric shock. Scientists have discovered how the timing of training sessions affects learning and have identified proteins and other molecules that strengthen synapses so the neuronal response is greater the next time Aplysia is stimulated. Many of the molecules and processes identified in Aplysia’s learning are also involved in human learning.
The fruit fly Drosophila is also commonly used to study behavior, especially how genes control behavior. For example, variations in a gene called ‘foraging’ determine whether flies tend to roam around as they eat or sit in one place. Flies with mutations in another gene called ‘timeless’ don’t have normal circadian rhythms. Mutations have been identified that affect the full gamut of Drosophila behaviors — from aggression to courtship, as well as learning and memory. Many of the affected genes have correlates in humans.
Addiction presents one of the most pressing challenges in studying human behavior — and researchers are still figuring out how to better understand and treat it. Some lab animals like rats will consume alcohol and drugs even if accompanied by a bitter taste or foot shock. Scientists have uncovered changes in the brains of animals exhibiting such addiction-like behaviors that mirror changes seen in the brains of humans with addiction disorders. Interestingly, some breeds of rats are very likely to exhibit addiction and relapse behaviors while others are more resistant. By comparing the genetics of two breeds of rats with different predispositions to cocaine addiction, scientists identified genes that were differentially turned on or off in the two breeds; the study suggests that these genes, and their epigenetic regulation, play a role in susceptibility to addiction. This type of research helps scientists understand why some people are more prone to addiction or relapse and could suggest ways to identify people at risk.
Behavior is also studied directly in humans. Early mapping of human behaviors to specific brain regions was done by observing personality changes in people who had lost small regions of their brain due to injuries or surgeries. For example, people who have lost their frontal lobe often become inconsiderate and impulsive. Modern imaging techniques also help scientists to pair brain regions with certain behaviors. For example, imaging allows researchers to see certain brain areas “light up” when a person is shown human faces, but not when they see faces of other animals. These techniques are also useful to better understand brain disorders — such as identifying brain regions responsible for auditory hallucinations in schizophrenia.
Adapted from the 8th edition of Brain Facts by Susan Rojahn.
CONTENT PROVIDED BY
BrainFacts/SfN
References
Bao, W., Jia, H., Finnema, S., Cai, Z., Carson, R. E., & Huang, Y. H. (2017). PET Imaging for Early Detection of Alzheimer's Disease: From Pathologic to Physiologic Biomarkers. PET clinics, 12(3), 329–350. https://doi.org/10.1016/j.cpet.2017.03.001
Berman, M. G., Jonides, J., & Nee, D. E. (2006). Studying Mind and Brain with fMRI. Social cognitive and affective neuroscience, 1(2), 158–161. https://doi.org/10.1093/scan/nsl019
Bögershausen, N., & Wollnik, B. (2013). Unmasking Kabuki syndrome. Clinical genetics, 83(3), 201–211. https://doi.org/10.1111/cge.12051
Boyden E. S. (2015). Optogenetics and the Future of Neuroscience. Nature neuroscience, 18(9), 1200–1201. https://doi.org/10.1038/nn.4094
Caraci, F., Leggio, G. M., Salomone, S., & Drago, F. (2017). New Drugs in Psychiatry: Focus on New Pharmacological Targets. F1000Research, 6, 397. https://doi.org/10.12688/f1000research.10233.1
Carter M. and Shieh J. C. (2015). Guide to Research Techniques in Neuroscience. Academic Press. p 164.
Carter N. P. (2007). Methods and Strategies for Analyzing Copy Number Variation Using DNA Microarrays. Nature genetics, 39(7 Suppl), S16–S21. https://doi.org/10.1038/ng2028
Chefer, V. I., Thompson, A. C., Zapata, A., & Shippenberg, T. S. (2009). Overview of Brain Microdialysis. Current protocols in neuroscience, Chapter 7, Unit 7.1. https://doi.org/10.1002/0471142301.ns0701s47
Clancy, S. (2008). Copy Number Variation. Nature Education, 1(1):95. https://www.nature.com/scitable/topicpage/copy-number-variation-445/
Cohen M. X. (2017). Where Does EEG Come From and What Does It Mean?. Trends in neurosciences, 40(4), 208–218. https://doi.org/10.1016/j.tins.2017.02.004
Courtney, K. E., & Ray, L. A. (2014). Methamphetamine: An Update on Epidemiology, Pharmacology, Clinical Phenomenology, and Treatment Literature. Drug and alcohol dependence, 143, 11–21. https://doi.org/10.1016/j.drugalcdep.2014.08.003
Cui, X., Bray, S., Bryant, D. M., Glover, G. H., & Reiss, A. L. (2011). A Quantitative Comparison of NIRS and fMRI Across Multiple Cognitive Tasks. NeuroImage, 54(4), 2808–2821. https://doi.org/10.1016/j.neuroimage.2010.10.069
Flagel, S. B., Chaudhury, S., Waselus, M., Kelly, R., Sewani, S., Clinton, S. M., Thompson, R. C., Watson, S. J., Jr, & Akil, H. (2016). Genetic Background and Epigenetic Modifications in the Core of the Nucleus Accumbens Predict Addiction-like Behavior in a Rat Model. Proceedings of the National Academy of Sciences of the United States of America, 113(20), E2861–E2870. https://doi.org/10.1073/pnas.1520491113
Gratten, J., Wray, N. R., Keller, M. C., & Visscher, P. M. (2014). Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nature neuroscience, 17(6), 782–790. https://doi.org/10.1038/nn.3708
Hämäläinen, M., Hari, R., Ilmoniemi, R. J., Knuutila, J., & Lounasmaa, O. V. (1993). Magnetoencephalography—Theory, Instrumentation, and Applications to Noninvasive Studies of the Working Human Brain. Reviews of modern Physics, 65(2), 413. https://journals.aps.org/rmp/abstract/10.1103/RevModPhys.65.413
Hanrieder, J., Phan, N. T., Kurczy, M. E., & Ewing, A. G. (2013). Imaging Mass Spectrometry in Neuroscience. ACS chemical neuroscience, 4(5), 666–679. https://doi.org/10.1021/cn400053c
Heather, J. M., & Chain, B. (2016). The Sequence of Sequencers: The History of Sequencing DNA. Genomics, 107(1), 1–8. https://doi.org/10.1016/j.ygeno.2015.11.003
Heidenreich, M., & Zhang, F. (2016). Applications of CRISPR-Cas Systems in Neuroscience. Nature reviews. Neuroscience, 17(1), 36–44. https://doi.org/10.1038/nrn.2015.2
Herbst, S. M., Proepper, C. R., Geis, T., Borggraefe, I., Hahn, A., Debus, O., Haeussler, M., von Gersdorff, G., Kurlemann, G., Ensslen, M., Beaud, N., Budde, J., Gilbert, M., Heiming, R., Morgner, R., Philippi, H., Ross, S., Strobl-Wildemann, G., Muelleder, K., Vosschulte, P., … Hehr, U. (2016). LIS1-associated Classic Lissencephaly: A Retrospective, Multicenter Survey of the Epileptogenic Phenotype and Response to Antiepileptic Drugs. Brain & development, 38(4), 399–406. https://doi.org/10.1016/j.braindev.2015.10.001
Hopf, F. W., & Lesscher, H. M. (2014). Rodent Models for Compulsive Alcohol Intake. Alcohol (Fayetteville, N.Y.), 48(3), 253–264. https://doi.org/10.1016/j.alcohol.2014.03.001
Johnson, A. C., & Greenwood-Van Meerveld, B. (2016). The Pharmacology of Visceral Pain. Advances in pharmacology (San Diego, Calif.), 75, 273–301. https://doi.org/10.1016/bs.apha.2015.11.002
Kandel, E. R., Dudai, Y., & Mayford, M. R. (2014). The Molecular and Systems Biology of Memory. Cell, 157(1), 163–186. https://doi.org/10.1016/j.cell.2014.03.001
Lee, G. J., Park, J. H., & Park, H. K. (2008). Microdialysis Applications in Neuroscience. Neurological research, 30(7), 661–668. https://doi.org/10.1179/174313208X289570
Leroy, A., Foucher, J. R., Pins, D., Delmaire, C., Thomas, P., Roser, M. M., Lefebvre, S., Amad, A., Fovet, T., Jaafari, N., & Jardri, R. (2017). fMRI Capture of Auditory Hallucinations: Validation of the Two-Steps Method. Human brain mapping, 38(10), 4966–4979. https://doi.org/10.1002/hbm.23707
Liu, Z., Ding, L., & He, B. (2006). Integration of EEG/MEG with MRI and fMRI. IEEE engineering in medicine and biology magazine : the quarterly magazine of the Engineering in Medicine & Biology Society, 25(4), 46–53. https://doi.org/10.1109/memb.2006.1657787
Lodish H., Berk A., Zipurksy S. L. et. al., editors. (2000). Molecular Cell Biology, 4th edition. Freeman, p 94, 140, 147-148, 268-269.
Malik, A. N., Vierbuchen, T., Hemberg, M., Rubin, A. A., Ling, E., Couch, C. H., Stroud, H., Spiegel, I., Farh, K. K., Harmin, D. A., & Greenberg, M. E. (2014). Genome-wide Identification and Characterization of Functional Neuronal Activity-Dependent Enhancers. Nature neuroscience, 17(10), 1330–1339. https://doi.org/10.1038/nn.3808
Mayford, M., Siegelbaum, S. A., & Kandel, E. R. (2012). Synapses and Memory Storage. Cold Spring Harbor perspectives in biology, 4(6), a005751. https://doi.org/10.1101/cshperspect.a005751
Maze, I., Shen, L., Zhang, B., Garcia, B. A., Shao, N., Mitchell, A., Sun, H., Akbarian, S., Allis, C. D., & Nestler, E. J. (2014). Analytical Tools and Current Challenges in the Modern Era of Neuroepigenomics. Nature neuroscience, 17(11), 1476–1490. https://doi.org/10.1038/nn.3816
National Human Genome Research Institute. (July 2017). An Overview of the Human Genome Project. Accessed July 17, 2017 at https://www.genome.gov/12011238/an-overview-of-the-human-genome-project/
National Institute of Mental Health. (2017). Brain Stimulation Therapies. Accessed July 17, 2017 at https://www.nimh.nih.gov/health/topics/brain-stimulation-therapies/brain-stimulation-therapies.shtml
Olgiati, S., Quadri, M., & Bonifati, V. (2016). Genetics of Movement Disorders in the Next-Generation Sequencing Era. Movement disorders, 31(4), 458–470. https://doi.org/10.1002/mds.26521
Perry, R. H., Blessed, G., Perry, E. K., & Tomlinson, B. E. (1980). Histochemical Observations on Cholinesterase Activities in the Brains of Elderly Normal and Demented (Alzheimer-type) Patients. Age and ageing, 9(1), 9–16. https://doi.org/10.1093/ageing/9.1.9
Purves D, Augustine GJ, Fitzpatrick D, et al., editors. (2008). Neuroscience. 4th edition. Sinauer Associates, Inc. p 3-5, 16-17, 19-21, 25-27, 181-187, 465, 559, 673-674, 715-717.
Sejnowski, T. J., Koch, C., & Churchland, P. S. (1988). Computational Neuroscience. Science (New York, N.Y.), 241(4871), 1299–1306. https://doi.org/10.1126/science.3045969
Sokolowski M. B. (2001). Drosophila: Genetics Meets Behaviour. Nature reviews. Genetics, 2(11), 879–890. https://doi.org/10.1038/35098592
Svoboda, K., & Yasuda, R. (2006). Principles of Two-Photon Excitation Microscopy and its Applications to Neuroscience. Neuron, 50(6), 823–839. https://doi.org/10.1016/j.neuron.2006.05.019
Turek, F. W., Pinto, L. H., Vitaterna, M. H., Penev, P. D., Zee, P. C., & Takahashi, J. S. (1995). Pharmacological and Genetic Approaches for the Study of Circadian Rhythms in Mammals. Frontiers in neuroendocrinology, 16(3), 191–223. https://doi.org/10.1006/frne.1995.1007
US National Library of Medicine, National Institutes of Health. (2017). Genetics Home Reference – Huntington Disease. Accessed July 17, 2017 at https://ghr.nlm.nih.gov/condition/huntington-disease#genes
Usdin, K., & Kumari, D. (2015). Repeat-mediated Epigenetic Dysregulation of the FMR1 Gene in the Fragile X-related Disorders. Frontiers in genetics, 6, 192. https://doi.org/10.3389/fgene.2015.00192
Yoshino, K., Oka, N., Yamamoto, K., Takahashi, H., & Kato, T. (2013). Functional Brain Imaging Using Near-infrared Spectroscopy During Actual Driving on an Expressway. Frontiers in human neuroscience, 7, 882. https://doi.org/10.3389/fnhum.2013.00882
What to Read Next
Also In Tools & Techniques
Trending
Popular articles on BrainFacts.org